XenoCell
- Project status: closed
Abstract
Background
Single-cell sequencing technologies provide unprecedented opportunities to deconvolve the genomic, transcriptomic or epigenomic heterogeneity of complex biological systems. Its application in samples from xenografts of patient-derived biopsies (PDX), however, is limited by the presence of cells originating from both the host and the graft in the analysed samples; in fact, in the bioinformatics workflows it is still a challenge discriminating between host and graft sequence reads obtained in a single-cell experiment.
Results
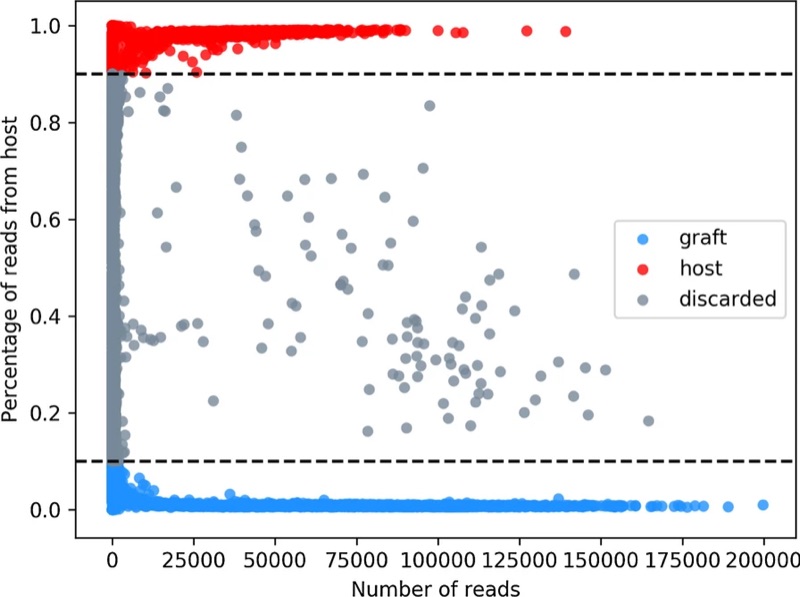
We have developed XenoCell, the first stand-alone pre-processing tool that performs fast and reliable classification of host and graft cellular barcodes from single-cell sequencing experiments. We show its application on a mixed species 50:50 cell line experiment from 10× Genomics platform, and on a publicly available PDX dataset obtained by Drop-Seq.
Conclusions
XenoCell accurately dissects sequence reads from any host and graft combination of species as well as from a broad range of single-cell experiments and platforms. It is open source and available at https://gitlab.com/XenoCell/XenoCell.
Citation
If you used XenoCell for your research, please cite the following publication:
Cheloni, S., Hillje, R., Luzi, L. et al. XenoCell: classification of cellular barcodes in single cell experiments from xenograft samples. BMC Med Genomics 14, 34 (2021). https://doi.org/10.1186/s12920-021-00872-8