Cerebro
- Project status: closed
Summary
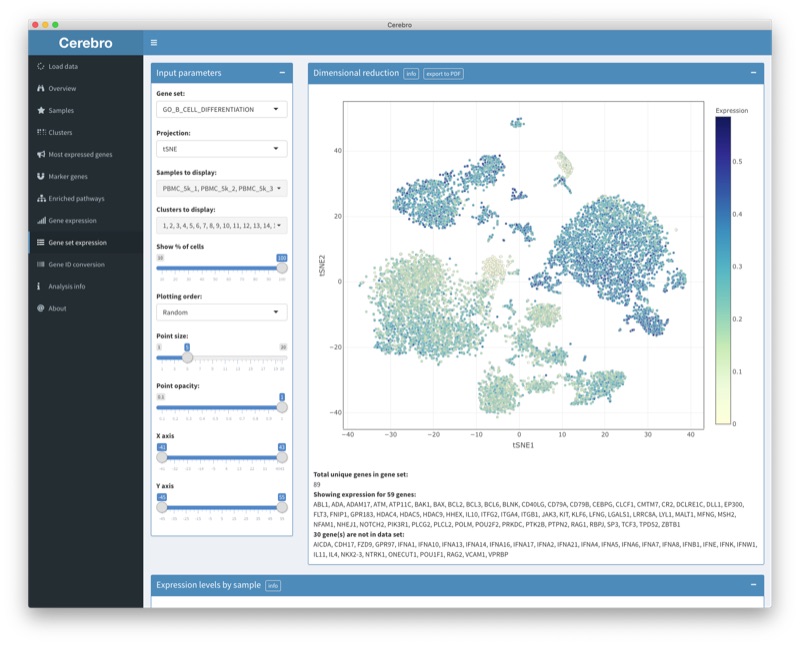
Transcriptomics data from single cells (scRNA-seq) are generated with unprecedented frequency due to steadily decreasing prices for kits and sequencing. Due to the often exploratory nature of scRNA-seq experiments, extensive knowledge of the biological system is required to properly infer cell types and derive conclusions. Even though public data sets are becoming more accessible through online platforms, direct interaction with their own data sets is often out of reach for experimental biologists. Cerebro is a Shiny- and Electron-based standalone desktop application for macOS and Windows which allows to investigate and visualize single cell transcriptomics data without any bioinformatic knowledge. Users have access to dimensional reductions, marker gene lists, results of functional analysis (gene ontology), and can retrieve expression of individual genes or gene lists. Overall, this tool aims to aid the study of transcriptomic data from single cells to ultimately facilitate scientific interpretation and discovery.
Additional material
- cerebroApp: R package that contains the Shiny application on which Cerebro is based
- Cerebro on GitHub: Standalone desktop application
- Poster about Cerebro I presented at the PhD Networking Days on January 9, 2020
Citation
If you used Cerebro for your research, please cite the following publication:
Roman Hillje, Pier Giuseppe Pelicci, Lucilla Luzi, Cerebro: interactive visualization of scRNA-seq data, Bioinformatics, btz877, https://doi.org/10.1093/bioinformatics/btz877