Distribution plot 5
Category: distribution
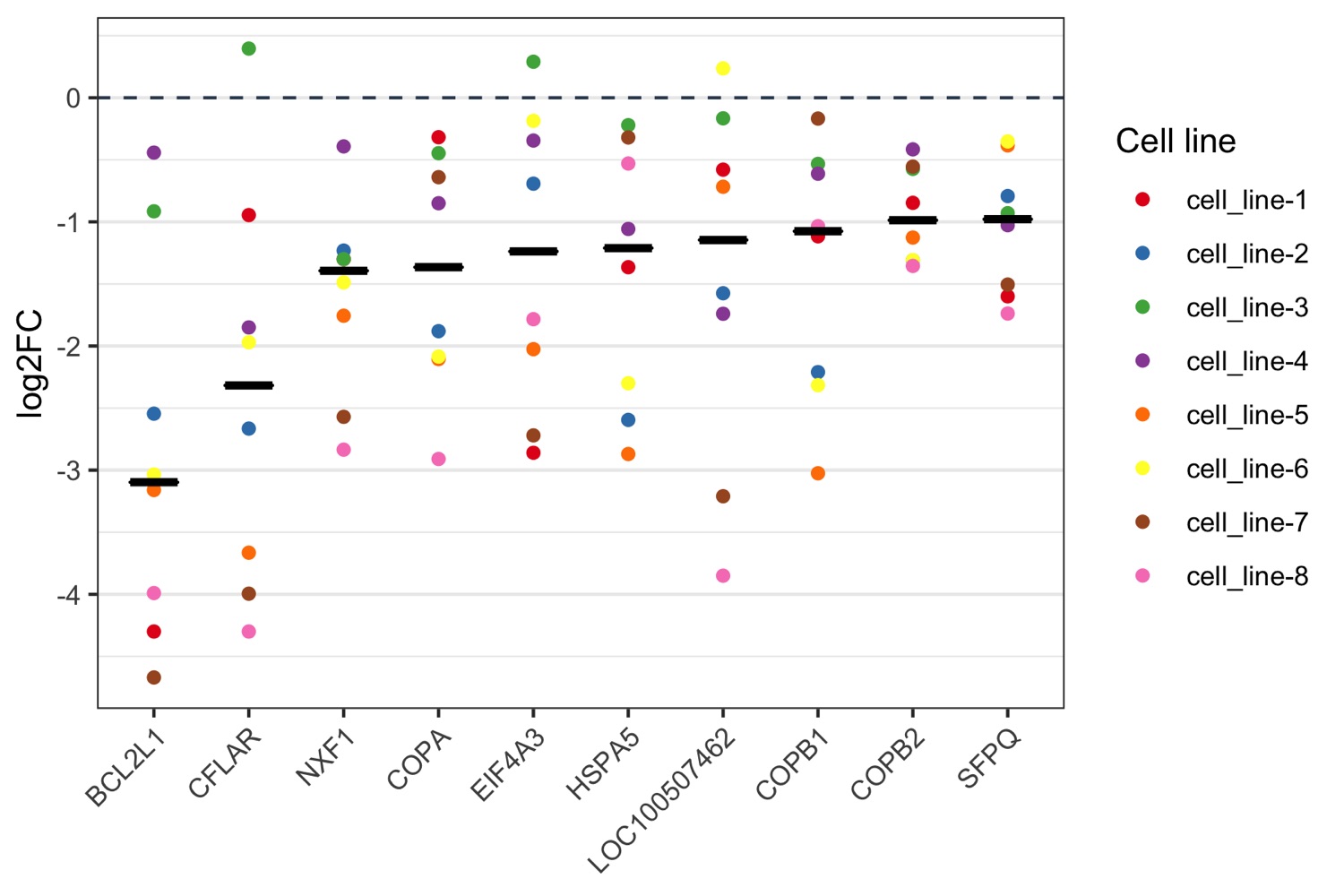
Data (CSV format):
,cell_line-1,cell_line-2,cell_line-3,cell_line-4,cell_line-5,cell_line-6,cell_line-7,cell_line-8
BCL2L1,-4.3,-2.545,-0.915,-0.442,-3.16,-3.035,-4.67,-3.99
CFLAR,-0.945,-2.665,0.396,-1.85,-3.665,-1.97,-3.995,-4.3
NXF1,-1.3,-1.231,-1.3,-0.392,-1.756,-1.488,-2.57,-2.835
COPA,-0.318,-1.88,-0.447,-0.85,-2.105,-2.085,-0.64,-2.91
EIF4A3,-2.86,-0.692,0.29,-0.345,-2.025,-0.186,-2.72,-1.784
HSPA5,-1.365,-2.595,-0.221,-1.057,-2.87,-2.3,-0.32,-0.53
LOC100507462,-0.579,-1.575,-0.166,-1.74,-0.717,0.236,-3.21,-3.85
COPB1,-1.116,-2.21,-0.533,-0.612,-3.025,-2.315,-0.168,-1.035
COPB2,-0.847,-1.31,-0.573,-0.416,-1.126,-1.308,-0.555,-1.355
SFPQ,-1.6,-0.792,-0.93,-1.027,-0.384,-0.352,-1.505,-1.738
Plot:
library(tidyverse)
data <- read_csv("data.csv") %>%
pivot_longer(cols=c(2:length(.)), names_to = "cell_line", values_to = "log2FC")
colnames(data)[1] = "gene"
gene_order <- data |> group_by(gene) |> summarise(median = median(log2FC)) |>
arrange(median) |> pull(gene)
data |>
mutate(gene = factor(gene, levels=gene_order)) |>
ggplot(aes(gene, log2FC, group=gene, color=cell_line)) +
geom_hline(yintercept = 0, linetype = "dashed", color = "#34495e") +
geom_point() +
stat_summary(fun = median, fun.min = median, fun.max = median,
geom = "crossbar", width = 0.5, show.legend = FALSE) +
scale_color_brewer(name = "Cell line", palette = "Set1") +
theme_bw() +
theme(
axis.title.x = element_blank(),
axis.text.x = element_text(angle = 45, hjust = 1),
panel.grid.major.x = element_blank()
)
ggsave("5.png", width = 6, height = 4)