Cases of coronavirus (ongoing)
Mar 13, 2020
Introduction
A colleague of mine (hey 👋 @lost_efano) showed me this R package coronavirus which collects data about the virus.
I simply couldn’t resist to plot some of the data.
library(tidyverse)
library(coronavirus)
## change some country names in the coronavirus data
coronavirus_tmp <- coronavirus %>%
filter(type == 'confirmed') %>%
rename(country = Country.Region) %>%
select(date,country,cases) %>%
mutate(
country = ifelse(grepl(country, pattern = '^Mainland China$'), 'China', country),
country = ifelse(grepl(country, pattern = '^North Macedonia$'), 'Macedonia', country),
country = ifelse(grepl(country, pattern = '^US$'), 'United States', country),
country = ifelse(grepl(country, pattern = '^UK$'), 'United Kingdom', country)
)
## create vector of unique countries and dates
countries <- unique(coronavirus_tmp$country) %>% sort()
dates <- unique(coronavirus_tmp$date) %>% sort()
## assign 0 cases to all countries without record
zero_cases <- tibble(
country = countries,
cases = 0
)
## skeleton for summed up data
data_summed_up <- tibble(
date = character(),
country = character(),
code = character(),
cases = numeric()
)
## for every recoded date, sum up the number of cases up until then
for ( i in seq(length(dates)) ) {
temp_cases <- coronavirus_tmp %>%
filter(date <= dates[i]) %>%
bind_rows(zero_cases) %>%
group_by(country) %>%
summarize(cases = sum(cases)) %>%
mutate(date = as.character(dates[i]))
data_summed_up <- bind_rows(
data_summed_up,
temp_cases
)
}
## create log-scaled version of cases for plotting (colors look better)
data_summed_up <- data_summed_up %>%
mutate(
date = factor(date, levels = unique(date)),
cases_log = log10(cases+1)
)
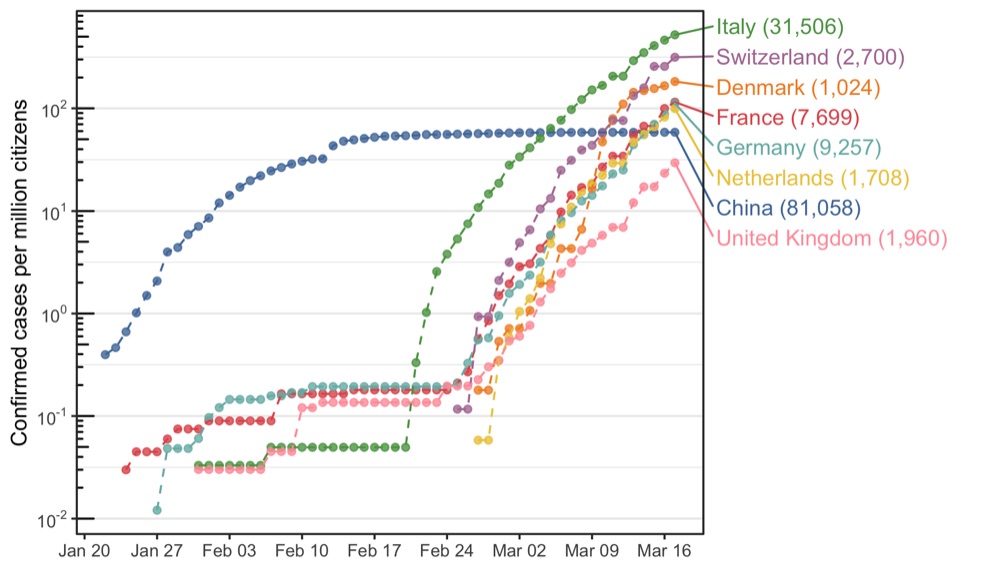
Highlight of specific countries
With labels
This plot wouldn’t have been possible without the help of these guys: https://stackoverflow.com/questions/54191372/log-scale-minor-tick-marks-on-outside-of-the-axis-line-with-annotation-logticks
The annotation_logticks() and coord_cartesian(clip = 'off') options results in log ticks outside of the plot which were removed by drawing a rectangle on top of them. Ingenious.
library(tidyverse)
library(coronavirus)
library(ggrepel)
library(ggthemes)
library(grid)
population_sizes <- tribble(
~country, ~population_size,
'Italy', 60.48,
'France', 66.89,
'Germany', 82.79,
'Switzerland', 8.57,
'United Kingdom', 66.44,
'Netherlands', 17.18,
'Denmark', 5.6,
'China', 1386
)
data_to_plot <- data_summed_up %>%
filter(
country %in% c('Italy','France','Germany','Switzerland','United Kingdom','Netherlands','Denmark','China'),
cases > 0
) %>%
mutate(date = as.Date(as.character(date))) %>%
left_join(population_sizes, by = 'country') %>%
mutate(cases_per_million = cases / population_size)
p <- data_to_plot %>%
ggplot(aes(date, cases_per_million, color = country)) +
geom_point(alpha = 0.8, size = 1.5) +
geom_line(linetype = 'dashed', show.legend = FALSE) +
scale_x_date(breaks = scales::pretty_breaks(n = 10)) +
scale_y_log10(
name = 'Confirmed cases per million citizens',
breaks = scales::trans_breaks('log10', function(x) 10^x),
labels = scales::trans_format('log10', scales::math_format(10^.x))
) +
annotation_logticks(sides ='l') +
coord_cartesian(clip = 'off') +
scale_color_tableau(name = '', guide = guide_legend(override.aes = list(size = 3))) +
geom_text_repel(
data = data_to_plot %>% filter(date == '2020-03-17'),
aes(
y = cases_per_million,
label = paste0(country, ' (', format(cases, big.mark = ',', trim = TRUE), ')')
),
x = as.Date('2020-03-17'), hjust = 0, direction = 'y', nudge_x = 4,
xlim = as.Date(c("2020-01-22", "2020-04-15")),
) +
theme_bw() +
theme(
legend.position = 'none',
axis.title.x = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank(),
plot.margin = unit(c(5.5,150,5.5,5.5), 'pt')
) +
annotation_custom(
grob = rectGrob(gp = gpar(col = NA)),
xmin = -100, xmax = min(data_to_plot$date),
ymin = -100, ymax = log10(min(data_to_plot$cases_per_million))-0.1
) +
annotation_custom(
grob = rectGrob(gp = gpar(col = NA)),
xmin = -100, xmax = min(data_to_plot$date),
ymin = log10(max(data_to_plot$cases_per_million))+0.2, ymax = 1000
)
ggsave('cases_per_million_citizens_with_labels.png', p, height = 4, width = 7)
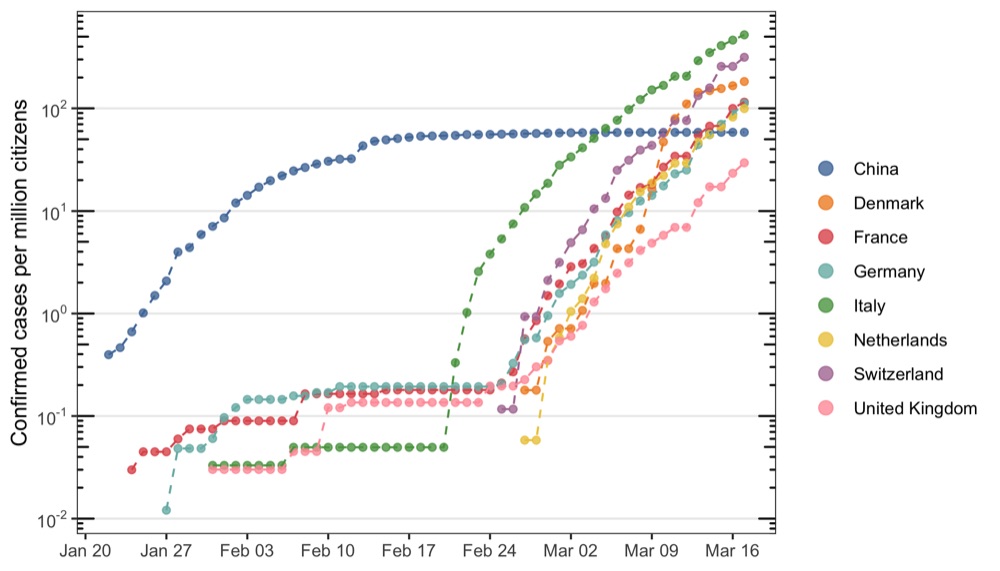
Without labels
library(tidyverse)
library(coronavirus)
library(ggrepel)
library(ggthemes)
population_sizes <- tribble(
~country, ~population_size,
'Italy', 60.48,
'France', 66.89,
'Germany', 82.79,
'Switzerland', 8.57,
'United Kingdom', 66.44,
'Netherlands', 17.18,
'Denmark', 5.6,
'China', 1386
)
data_to_plot <- data_summed_up %>%
filter(
country %in% c('Italy','France','Germany','Switzerland','United Kingdom','Netherlands','Denmark','China'),
cases > 0
) %>%
mutate(date = as.Date(as.character(date))) %>%
left_join(population_sizes, by = 'country') %>%
mutate(cases_per_million = cases / population_size)
p <- ggplot(data_to_plot, aes(date, cases_per_million, color = country)) +
geom_point(alpha = 0.8, size = 1.5) +
geom_line(linetype = 'dashed', show.legend = FALSE) +
scale_x_date(breaks = scales::pretty_breaks(n = 10)) +
scale_y_log10(
name = 'Confirmed cases per million citizens',
breaks = scales::trans_breaks('log10', function(x) 10^x),
labels = scales::trans_format('log10', scales::math_format(10^.x))
) +
annotation_logticks(sides ='lr') +
scale_color_tableau(name = '', guide = guide_legend(override.aes = list(size = 3))) +
theme_bw() +
theme(
legend.position = 'right',
axis.title.x = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor = element_blank()
)
ggsave('cases_per_million_citizens.png', p, height = 4, width = 7)
Animated
library(tidyverse)
library(coronavirus)
library(ggthemes)
library(gganimate)
population_sizes <- tribble(
~country, ~population_size,
'Italy', 60.48,
'France', 66.89,
'Germany', 82.79,
'Switzerland', 8.57,
'United Kingdom', 66.44,
'Netherlands', 17.18,
'Denmark', 5.6,
'China', 1386
)
data_to_plot <- data_summed_up %>%
filter(
country %in% c('Italy','France','Germany','Switzerland','United Kingdom','Netherlands','Denmark','China'),
cases > 0
) %>%
mutate(date = as.Date(as.character(date))) %>%
left_join(population_sizes, by = 'country') %>%
mutate(cases_per_million = cases / population_size)
p <- data_to_plot %>%
ggplot(aes(date, cases_per_million, color = country)) +
geom_point(alpha = 0.8, size = 1.5) +
geom_line(linetype = 'dashed', show.legend = FALSE) +
geom_segment(
aes(yend = cases_per_million), xend = as.Date('2020-03-20'), linetype = 2
) +
geom_text(
aes(label = paste0(country, ' (', format(cases, big.mark = ',', trim = TRUE), ')')),
x = as.Date('2020-03-21'), hjust = 0
) +
scale_x_date(breaks = scales::pretty_breaks(n = 10)) +
scale_y_log10(
name = 'Confirmed cases per million citizens',
breaks = scales::trans_breaks('log10', function(x) 10^x),
labels = scales::trans_format('log10', scales::math_format(10^.x))
) +
coord_cartesian(clip = 'off') +
scale_color_tableau(guide = guide_legend(override.aes = list(size = 3))) +
labs(title = 'Confirmed cases per million citizens (total cases in brackets)') +
theme_bw() +
theme(
legend.position = 'none',
axis.title.x = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank(),
plot.margin = unit(c(5.5, 140, 5.5, 5.5), 'pt')
) +
transition_reveal(date) +
ease_aes('linear')
animation <- animate(p, height = 400, width = 600, end_pause = 30)
anim_save('cases_per_million_citizens_with_labels.gif', animation)
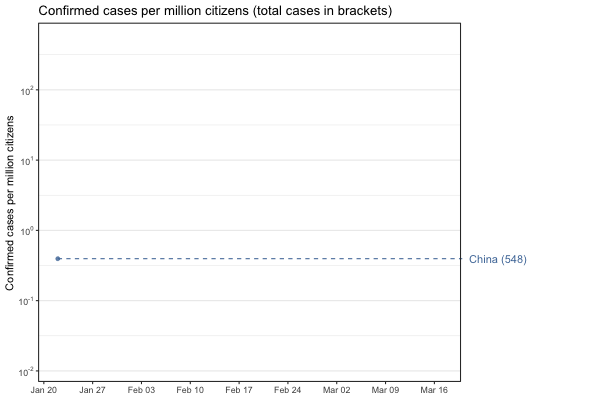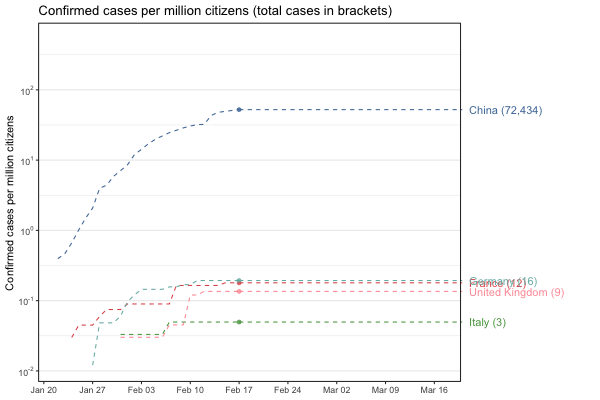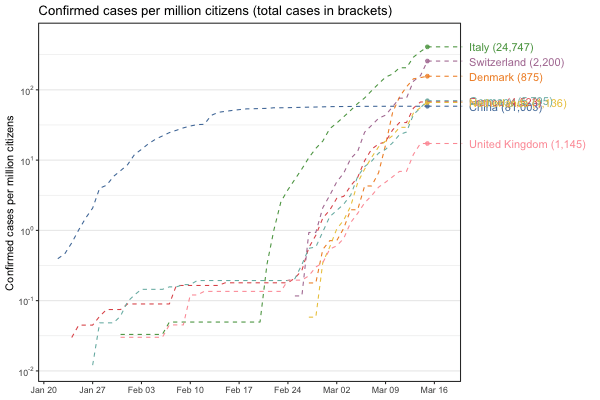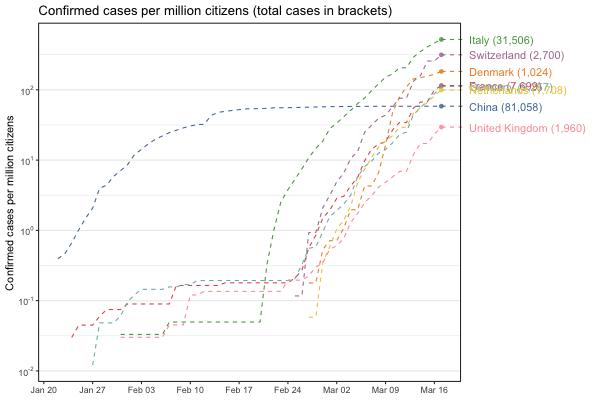
Interactive world map
For now, I made an animated world map showing the number of cases by country since 22.01.2020.
library(tidyverse)
library(coronavirus)
library(plotly)
## download country codes
country_codes <- read.csv('https://raw.githubusercontent.com/plotly/datasets/master/2014_world_gdp_with_codes.csv') %>%
select(COUNTRY, CODE) %>%
rename(country = COUNTRY) %>%
mutate(
country = as.character(country),
country = ifelse(grepl(country, pattern = '^Korea, North$'), 'North Korea', country),
country = ifelse(grepl(country, pattern = '^Korea, South$'), 'South Korea', country)
)
## change some country names in the coronavirus data
coronavirus_tmp <- coronavirus %>%
filter(type == 'confirmed') %>%
rename(country = Country.Region) %>%
select(date,country,cases) %>%
mutate(
country = ifelse(grepl(country, pattern = '^Mainland China$'), 'China', country),
country = ifelse(grepl(country, pattern = '^North Macedonia$'), 'Macedonia', country),
country = ifelse(grepl(country, pattern = '^US$'), 'United States', country),
country = ifelse(grepl(country, pattern = '^UK$'), 'United Kingdom', country)
)
## create vector of unique countries and dates
countries <- unique(country_codes$country) %>% sort()
dates <- unique(coronavirus_tmp$date) %>% sort()
## assign 0 cases to all countries without record
zero_cases <- tibble(
country = countries,
cases = 0
)
## skeleton for summed up data
data_summed_up <- tibble(
date = character(),
country = character(),
code = character(),
cases = numeric()
)
## for every recoded date, sum up the number of cases up until then
for ( i in seq(length(dates)) ) {
temp_cases <- coronavirus_tmp %>%
filter(date <= dates[i]) %>%
bind_rows(zero_cases) %>%
group_by(country) %>%
summarize(cases = sum(cases)) %>%
mutate(date = as.character(dates[i]))
data_summed_up <- bind_rows(
data_summed_up,
temp_cases
)
}
## create log-scaled version of cases for plotting (colors look better)
## and add country codes
data_summed_up <- data_summed_up %>%
mutate(
date = factor(date, levels = unique(date)),
cases_log = log10(cases+1)
) %>%
full_join(country_codes, by = 'country')
## create plot
p <- plot_geo(data_summed_up) %>%
add_trace(
z = ~cases_log,
color = ~cases,
colors = 'Blues',
frame = ~date,
hoverinfo = 'text',
text = ~paste0(data_summed_up$country, ': ', format(data_summed_up$cases, trim = TRUE, big.mark = ',')),
locations = ~CODE,
marker = list(
line = list(
color = toRGB("grey"),
width = 0.5
)
),
showscale = FALSE
) %>%
layout(
geo = list(
showframe = FALSE,
showcoastlines = FALSE,
projection = list(type = 'winkel tripel')
)
)
htmlwidgets::saveWidget(p, 'world_map_animated.html')
Click the “Play” to start the animation or manually move the slider to see the date of interest.
Confirmed cases in China
library(tidyverse)
library(coronavirus)
library(patchwork)
p1 <- coronavirus %>%
filter(
type == 'confirmed',
grepl(Country.Region, pattern = 'China')
) %>%
group_by(date) %>%
summarize(cases = sum(cases)) %>%
mutate(cases_sum = cumsum(cases)) %>%
ggplot(aes(date, cases_sum)) +
geom_smooth(method = 'loess', formula = 'y ~ x', color = 'red') +
geom_point() +
labs(title = 'Total number of COVID-19 cases in China', subtitle = 'Linear scale') +
scale_x_date(breaks = scales::pretty_breaks(n = 10)) +
scale_y_continuous(name = 'Number of cases', labels = scales::comma) +
theme_bw() +
theme(
axis.title.x = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()
)
p2 <- coronavirus %>%
filter(
type == 'confirmed',
grepl(Country.Region, pattern = 'China')
) %>%
group_by(date) %>%
summarize(cases = sum(cases)) %>%
mutate(cases_sum = cumsum(cases)) %>%
ggplot(aes(date, cases_sum)) +
geom_smooth(method = 'loess', formula = 'y ~ x', color = 'red') +
geom_point() +
labs(subtitle = 'Log scale') +
scale_x_date(breaks = scales::pretty_breaks(n = 10)) +
scale_y_log10(name = 'Number of cases', labels = scales::comma) +
theme_bw() +
theme(
axis.title.x = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()
)
ggsave('total_cases_china.png', p1 + p2 + plot_layout(ncol = 1), height = 6, width = 8)
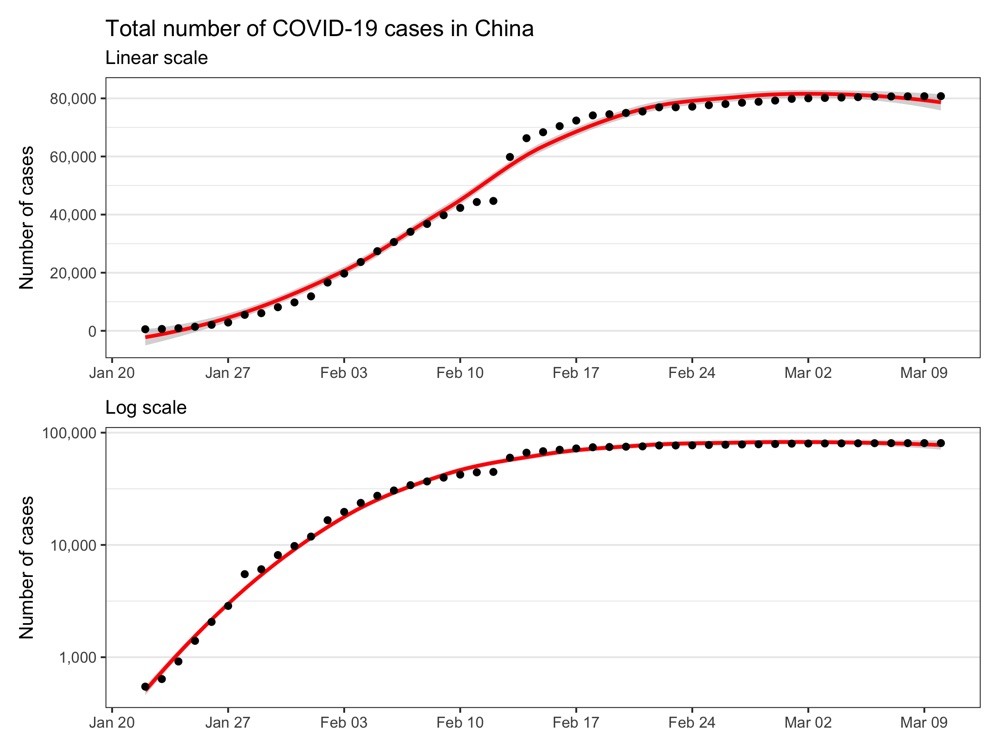
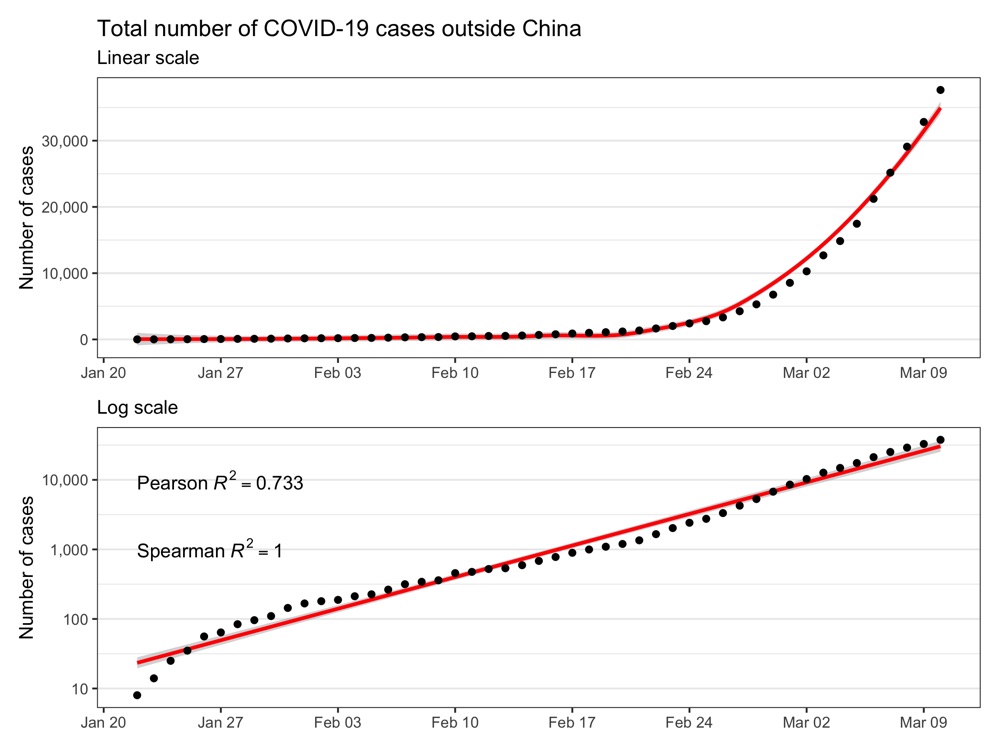
Confirmed cases outside of China
library(tidyverse)
library(coronavirus)
library(patchwork)
coronavirus_tmp <- coronavirus %>%
filter(
type == 'confirmed',
grepl(Country.Region, pattern = 'China') == FALSE
) %>%
rename(country = Country.Region) %>%
select(date,cases) %>%
group_by(date) %>%
summarize(cases = sum(cases)) %>%
mutate(cases_sum = cumsum(cases))
p1 <- ggplot(coronavirus_tmp, aes(date, cases_sum)) +
geom_smooth(method = 'loess', formula = 'y ~ x', color = 'red') +
geom_point() +
labs(title = 'Total number of COVID-19 cases outside China', subtitle = 'Linear scale') +
scale_x_date(breaks = scales::pretty_breaks(n = 10)) +
scale_y_continuous(name = 'Number of cases', labels = scales::comma) +
theme_bw() +
theme(
axis.title.x = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()
)
correlation_pearson <- cor(as.numeric(coronavirus_tmp$date), coronavirus_tmp$cases_sum, method = 'pearson')
correlation_spearman <- cor(as.numeric(coronavirus_tmp$date), coronavirus_tmp$cases_sum, method = 'spearman')
label <- list(
paste0('Pearson~italic(R)^2 == ', round(correlation_pearson, 3)),
paste0('Spearman~italic(R)^2 == ', round(correlation_spearman, 3))
)
p2 <- ggplot(coronavirus_tmp, aes(date, cases_sum)) +
geom_smooth(method = 'lm', formula = 'y ~ x', color = 'red') +
geom_point() +
labs(subtitle = 'Log scale') +
scale_x_date(breaks = scales::pretty_breaks(n = 10)) +
scale_y_log10(name = 'Number of cases', labels = scales::comma) +
annotate('text', x = coronavirus_tmp$date[1], y = c(1E4, 1E3), hjust = 0, parse = TRUE, label = label) +
theme_bw() +
theme(
axis.title.x = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()
)
ggsave('total_cases_outside_china.png', p1 + p2 + plot_layout(ncol = 1), height = 6, width = 8)
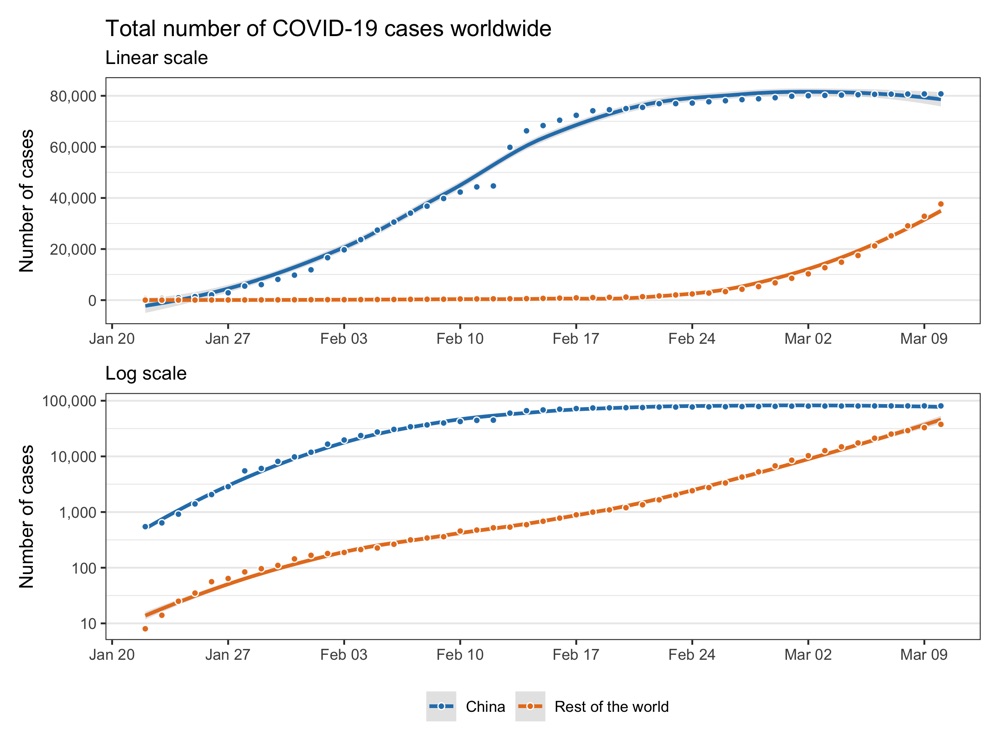
Confirmed cases worldwide
library(tidyverse)
library(coronavirus)
library(patchwork)
coronavirus_tmp <- coronavirus %>%
filter(type == 'confirmed') %>%
mutate(group = ifelse(grepl(Country.Region, pattern = 'China'), 'China', 'Rest of the world')) %>%
group_by(group,date) %>%
summarize(cases = sum(cases)) %>%
mutate(cases_sum = cumsum(cases))
colors_here <- c('#2980b9', '#e67e22')
p1 <- ggplot(coronavirus_tmp, aes(date, cases_sum, fill = group, color = group, group = group)) +
geom_smooth(fill = 'grey', method = 'loess', formula = 'y ~ x') +
geom_point(color = 'white', shape = 21) +
labs(title = 'Total number of COVID-19 cases worldwide', subtitle = 'Linear scale') +
scale_x_date(breaks = scales::pretty_breaks(n = 10)) +
scale_y_continuous(name = 'Number of cases', labels = scales::comma) +
scale_color_manual(name = '', values = colors_here) +
scale_fill_manual(name = '', values = colors_here) +
theme_bw() +
theme(
legend.position = 'none',
axis.title.x = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()
)
p2 <- ggplot(coronavirus_tmp, aes(date, cases_sum, fill = group, color = group, group = group)) +
geom_smooth(fill = 'grey', method = 'loess', formula = 'y ~ x') +
geom_point(color = 'white', shape = 21) +
labs(subtitle = 'Log scale') +
scale_x_date(breaks = scales::pretty_breaks(n = 10)) +
scale_y_log10(name = 'Number of cases', labels = scales::comma) +
scale_color_manual(name = '', values = colors_here) +
scale_fill_manual(name = '', values = colors_here) +
theme_bw() +
theme(
legend.position = 'bottom',
axis.title.x = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()
)
ggsave('total_cases_china_and_rest.png', p1 + p2 + plot_layout(ncol = 1), height = 6, width = 8)
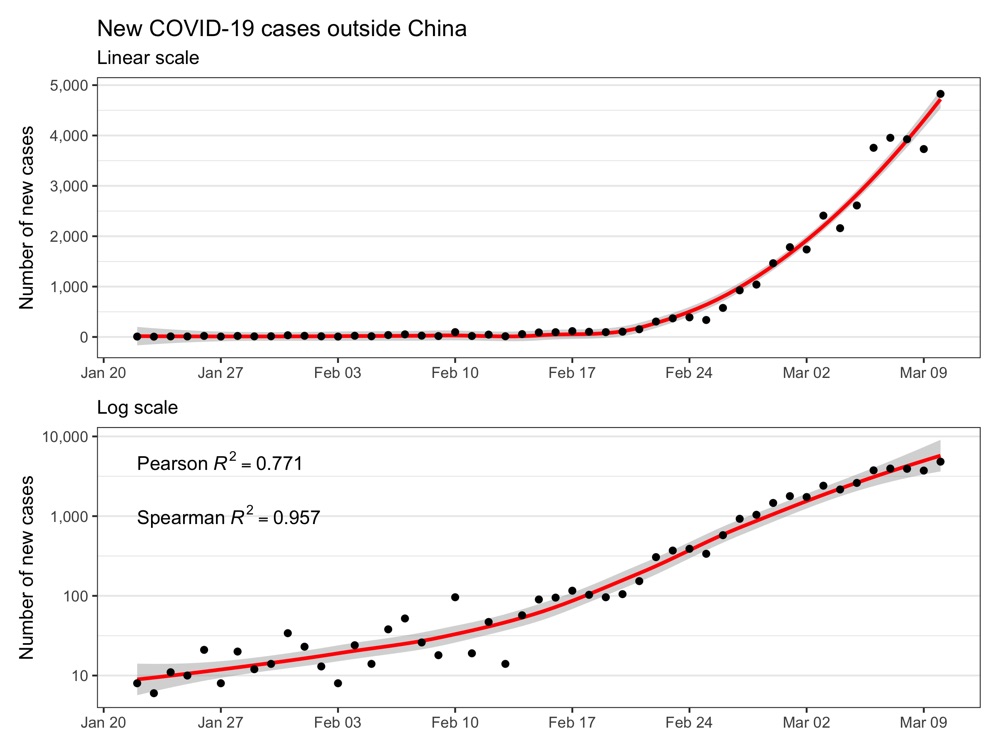
New cases outside of China
library(tidyverse)
library(coronavirus)
library(patchwork)
coronavirus_tmp <- coronavirus %>%
filter(
type == 'confirmed',
grepl(Country.Region, pattern = 'China') == FALSE
) %>%
group_by(date) %>%
summarize(cases = sum(cases))
p1 <- ggplot(coronavirus_tmp, aes(date, cases)) +
geom_smooth(method = 'loess', formula = 'y ~ x', color = 'red') +
geom_point() +
labs(title = 'New COVID-19 cases outside China', subtitle = 'Linear scale') +
scale_x_date(breaks = scales::pretty_breaks(n = 10)) +
scale_y_continuous(name = 'Number of new cases', labels = scales::comma) +
theme_bw() +
theme(
axis.title.x = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()
)
correlation_pearson <- cor(as.numeric(coronavirus_tmp$date), coronavirus_tmp$cases, method = 'pearson')
correlation_spearman <- cor(as.numeric(coronavirus_tmp$date), coronavirus_tmp$cases, method = 'spearman')
label <- list(
paste0('Pearson~italic(R)^2 == ', round(correlation_pearson, 3)),
paste0('Spearman~italic(R)^2 == ', round(correlation_spearman, 3))
)
p2 <- ggplot(coronavirus_tmp, aes(date, cases)) +
geom_smooth(method = 'loess', formula = 'y ~ x', color = 'red') +
geom_point() +
labs(subtitle = 'Log scale') +
scale_x_date(breaks = scales::pretty_breaks(n = 10)) +
scale_y_log10(name = 'Number of new cases', labels = scales::comma) +
annotate('text', x = coronavirus_tmp$date[1], y = c(5E3, 1E3), hjust = 0, parse = TRUE, label = label) +
theme_bw() +
theme(
axis.title.x = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()
)
ggsave('new_cases_outside_china.png', p1 + p2 + plot_layout(ncol = 1), height = 6, width = 8)